lecture 17: t-test and bootstrapping
2024-03-21
Recap
Goal of t-tests: Establish the difference of mean values between 2 samples
Reminder on sampling from the red and white balls pictures
Reminder on using R to do simulations
Today’s class: from sampling distribution -> p-values ; SEM ; CI
Reminder on hypothesis testing
We discussed example in lec13:
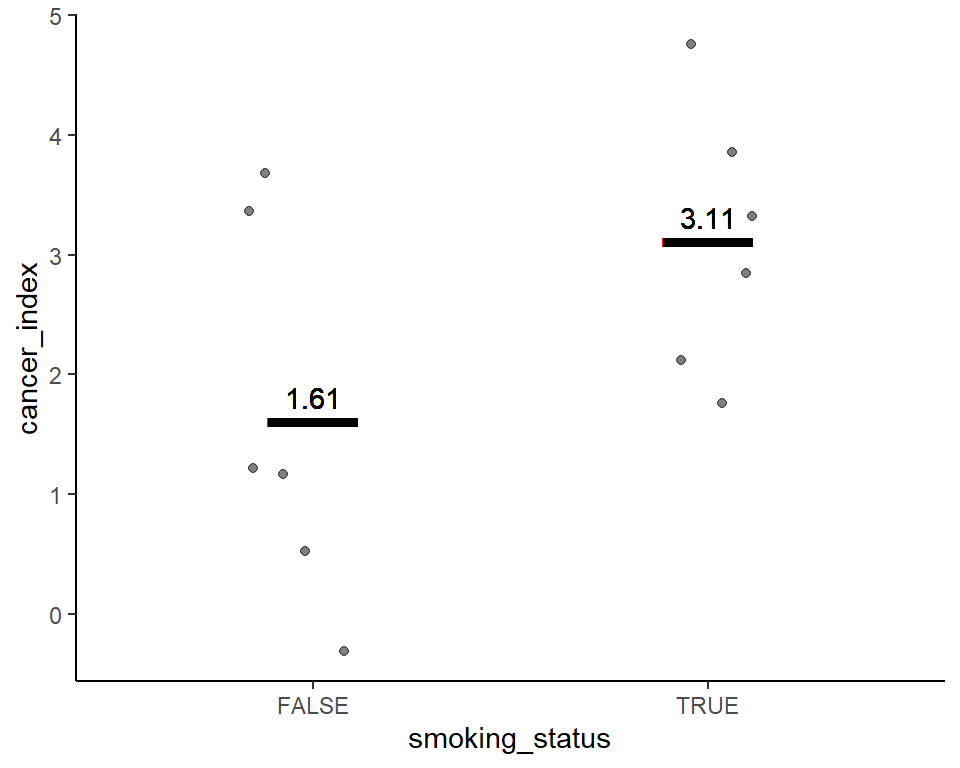
Smoking status is statistically significantly associated with higher cancer incidence
NULL hypothesis (for t-test) = Sample means are equal/ samples belong to the same population/distribution
Alternate hypothesis = (choose only 1 per t-test!)
Sample means are unequal (two tailed t-test)
Sample A’s mean > sample B’s mean (one tailed t-test)
Difference of mean between samples
2 samples = t-test
more samples = ANOVA
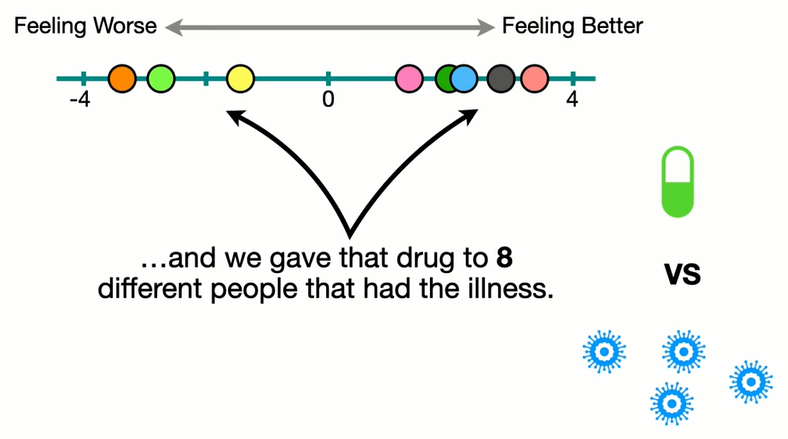
Sampling from a population
Population

Sample

Re-sampling = shuffling within the sample (w replacement)
Sample
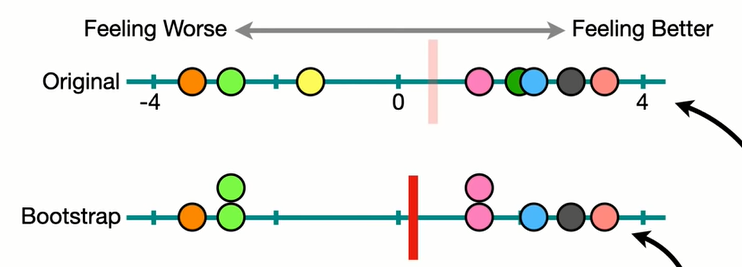
Bootstrapped sample (re-sampling, with replacement)
Reminder: Recipe for simulation/lec12
Key difference is how the randomness comes in
For simulation, we use
rnorm(normal dist, random number) /runif(uniform dist, rv) etc. to generate random numbers from different distributionsFor sampling, we use
sample(population_vector, size = sample_size, replace = FALSE)to select a random sample (subset) of the populationFor bootstrapping, we use
sample(sample_vector, size = sample_size, replace = TRUE)to select a random bootstrap sample from the sample
For repeating steps (iteration), you can use
for () loops: beginner friendlymap()like vectorized functions: succinct code, needs some head breaking to get used to
Today’s class
- Doing a t-test in R
- Using re-sampling distributions to get p-values, Standard Error of Mean (SEM) and confidence intervals (CI)
- How does this relate to a t-test?
- Watch the nice Youtube videos linked in the course schedule for lecture 17
Dataset for t-testing

Many flavours of t-tests
1-sample vs 2-sample
- >2 samples ~ do ANOVA
1-tailed vs 2-tailed
Paired vs unpaired
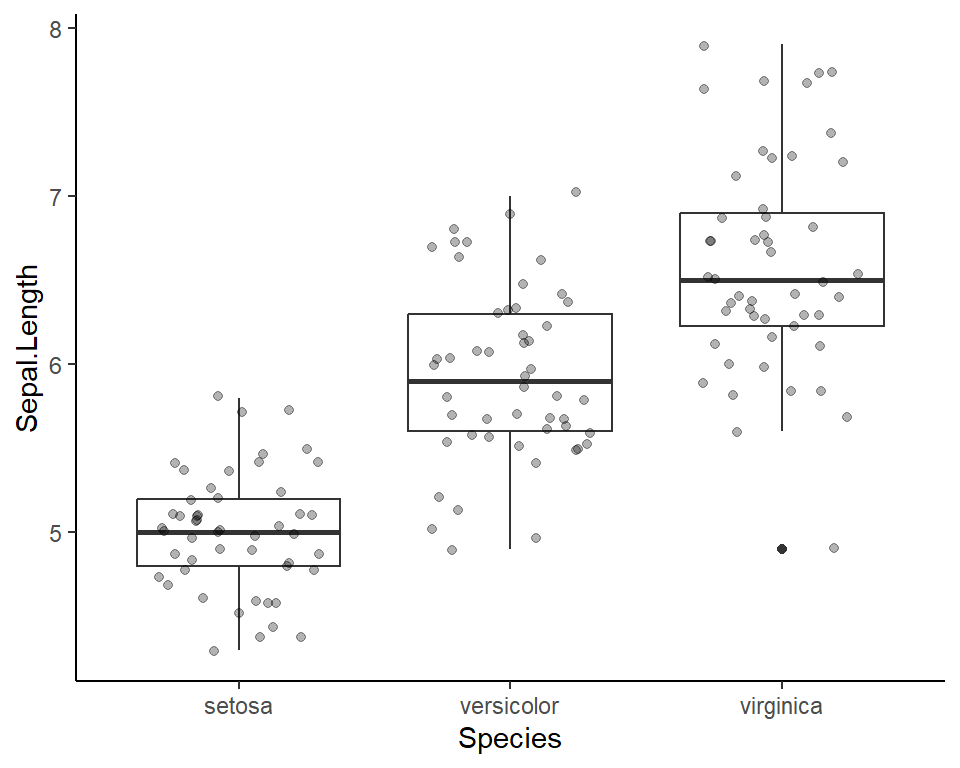
Visualize before 2-sample t-test
Here’s a plot outlining the data to use for t-test
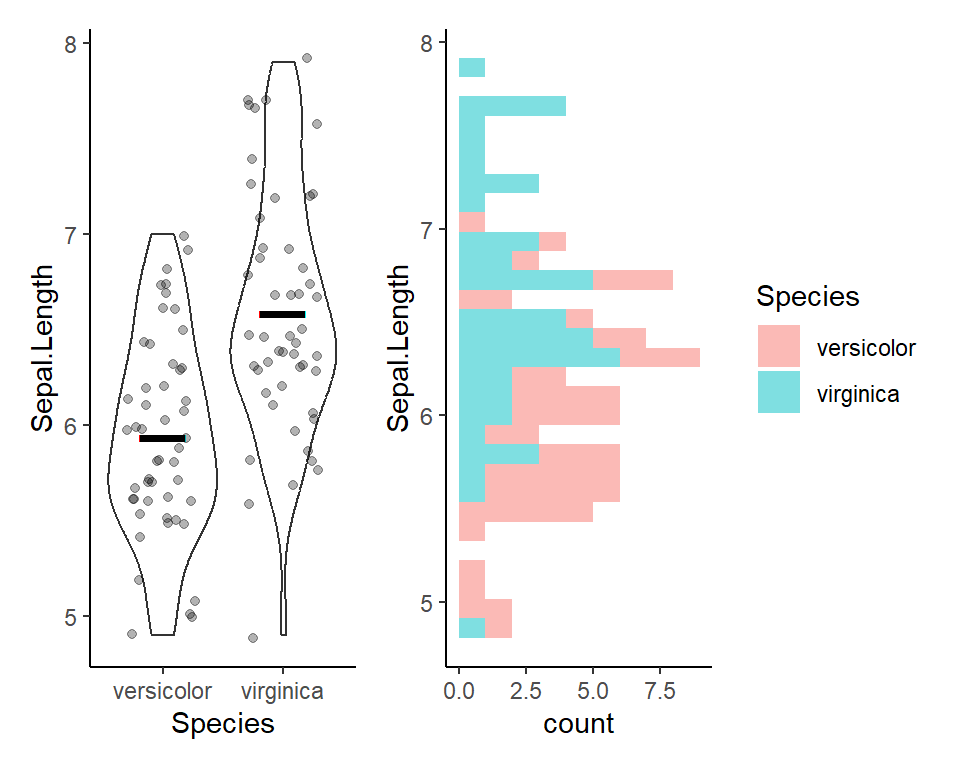
2-sample t-test in R
Welch Two Sample t-test
data: Sepal.Length by Species
t = -5.6292, df = 94.025, p-value = 1.866e-07
alternative hypothesis: true difference in means between group versicolor and group virginica is not equal to 0
95 percent confidence interval:
-0.8819731 -0.4220269
sample estimates:
mean in group versicolor mean in group virginica
5.936 6.588 What is p-value?
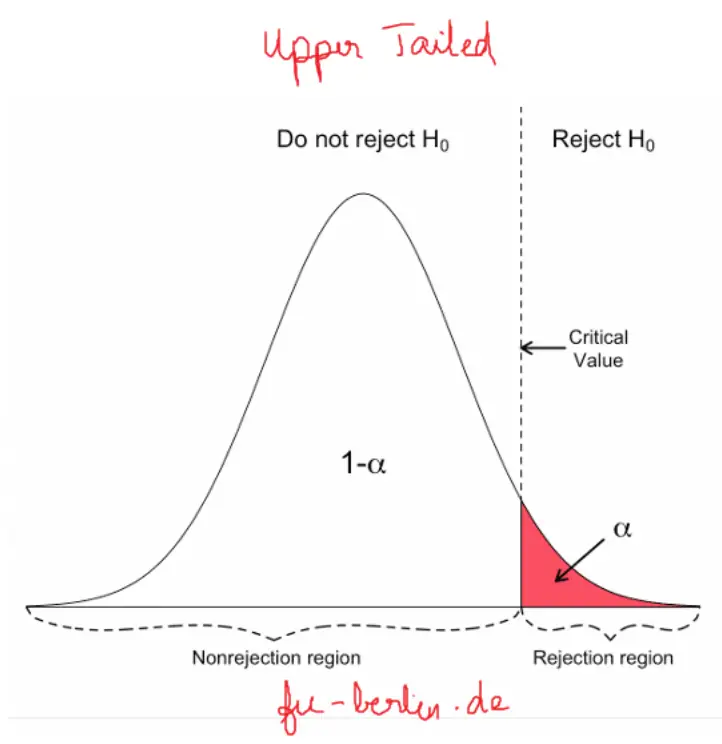
p-value is the probability that the observed difference of means (or more extreme) can occur by chance if the NULL hypothesis is TRUE
This is calculated by
plotting a t-distribution around the null hypothesis mean difference (typically 0)
mark the observed mean difference
Find the tail of the distribution beyond the observed value
Bootstrapping to understand t-test better
Bootstrapping shows us the variability around the mean, by virtually repeating the experiment a bunch of times
For 1 sample, this is how the bootstrapped distribution looks like
Code
[1] -0.85 -0.50 -0.34 -0.07 0.43 0.57 0.65 1.25 1.27 1.36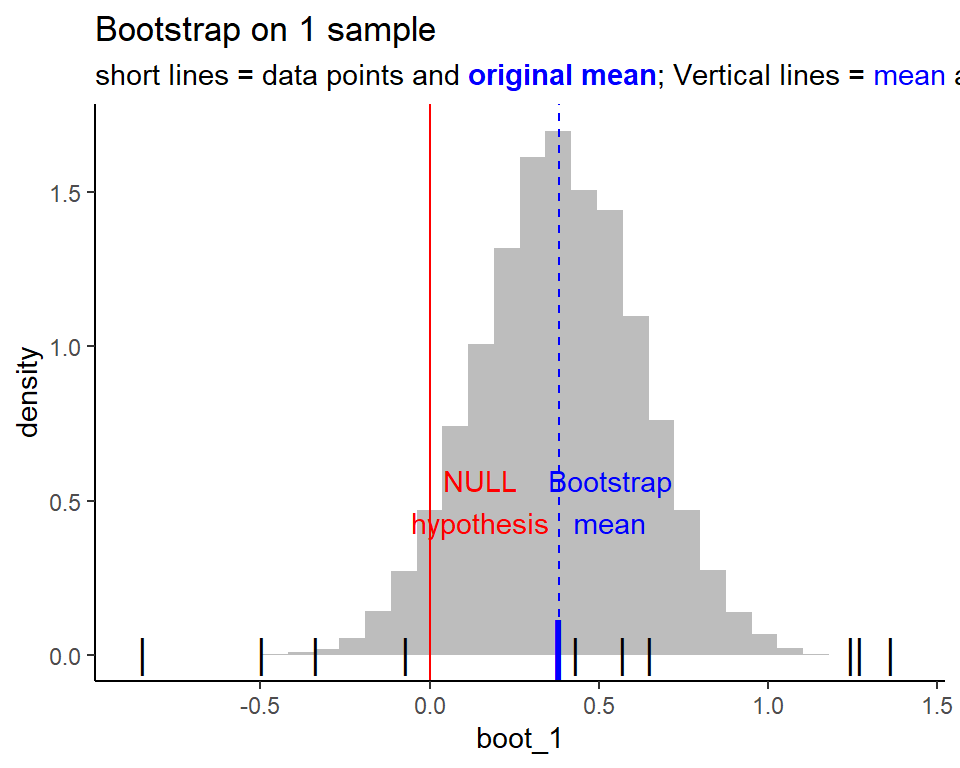
1-sample t-test
Null hypothesis (\(H_0\)) ~ [\(\mu = \mu_0\)] => the mean of the data is \(\mu_0\)
- We will test for: \(\mu = 0\) for this dataset
One Sample t-test
data: sample_1
t = 1.5179, df = 9, p-value = 0.1634
alternative hypothesis: true mean is not equal to 0
95 percent confidence interval:
-0.1857309 0.9433002
sample estimates:
mean of x
0.3787847 [1] "p-value for t-test is: 0.16"1-sample t-test w bootstrapping
To get p-value from the bootstrapping, we need to find the area of the tails. To facilitate understanding, we shift the distribution to fit the null hypothesis. Which means, the mean of the distribution should be moved to the null hypothesis!
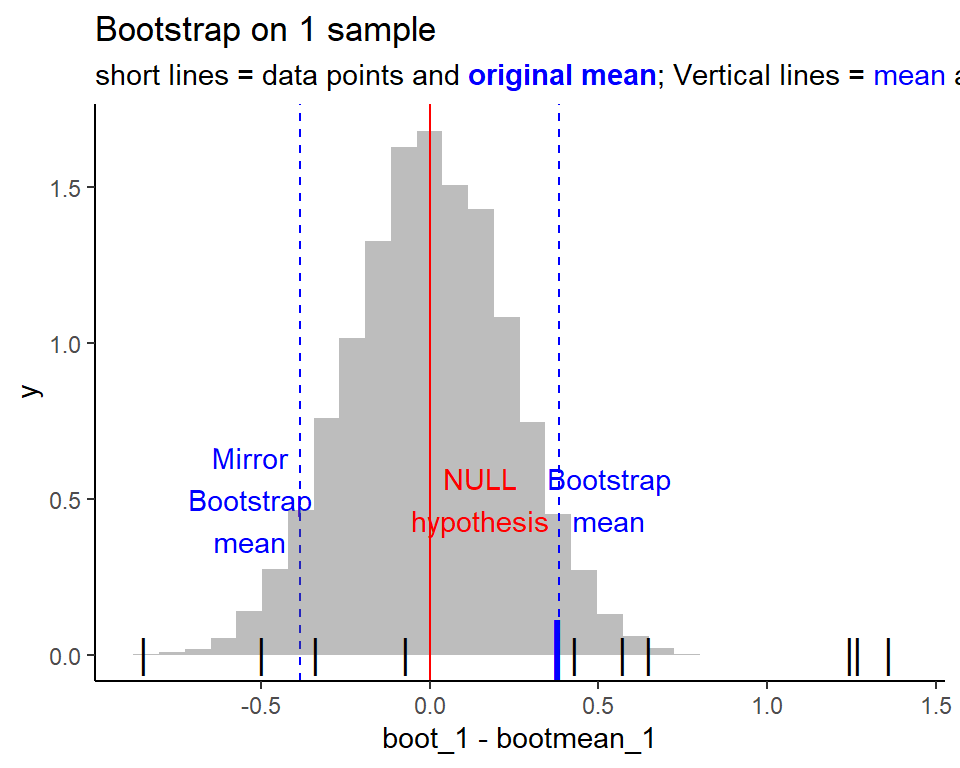
Showing the 2 tails
For a 2 tailed test, the p-value corresponds to the area under these two tails
Tail 1: \(\mu > \mu_0\)
Tail 2: \(\mu < \mu_0\)
Both tails: \(\mu != \mu_0\)
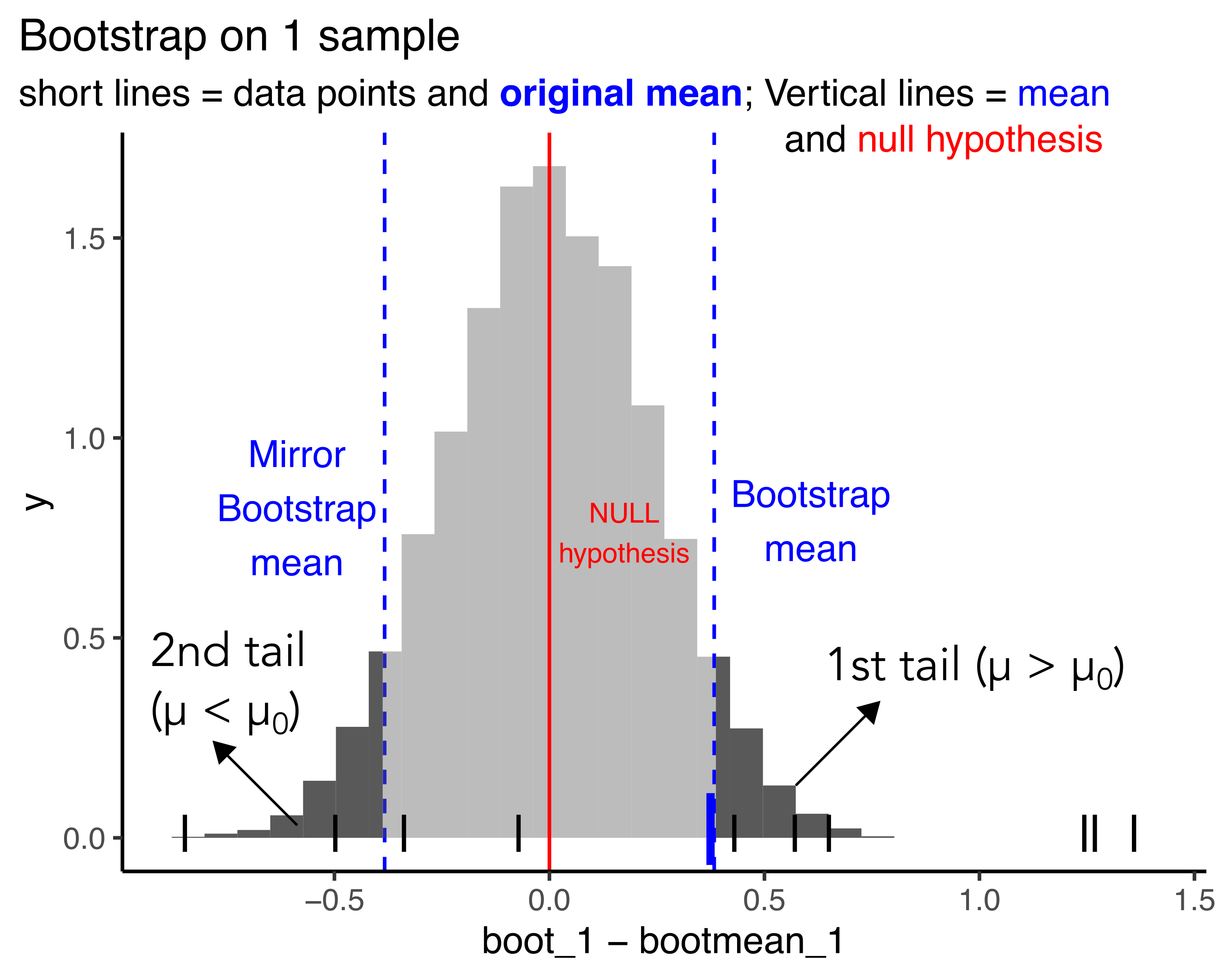
Calculating the p-value from bootstrap dist.
Graphically area is easy to visualize,
For calculation, probability is just the number of values in the tails / total number of values
first_tail <- sum(boot_1 - bootmean_1 > bootmean_1)
second_tail <- sum(boot_1 < 0)
boot_p_val <- (first_tail + second_tail)/length(boot_1)
str_c('p-value for bootstrapping t-test: ', boot_p_val)[1] "p-value for bootstrapping t-test: 0.1067"[1] "p-value for t-test is: 0.16"p-value for t-test is: 0.16
p-value for t-test is: 0.1067
Exploring SEM, CI within bootstrap dist.
(SEM) Standard error of mean = std deviation of the bootstrapping distribution
(CI) 95% confidence interval => 95% of the mean distribution area lies within this range
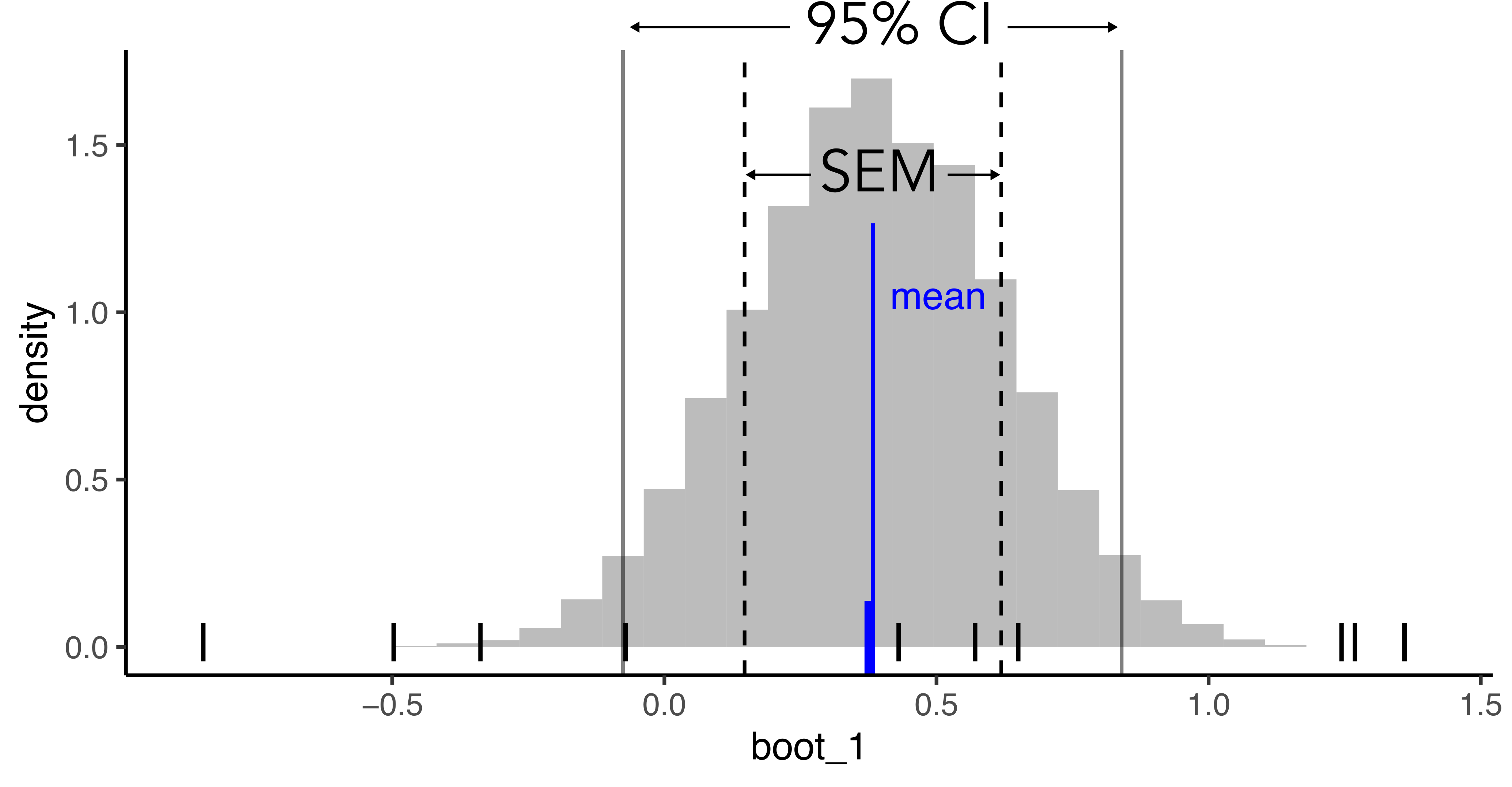
Onto the worksheet : 2 samples t-test
Please download/git pull the class17_t-test_bootstrapping.qmd worksheet for today from Github
- Use the viridis dataset : virginica vs versicolor
- [second section: Mutate new columns to generate: versicolor (+ 0.5)]
- Do t.test and record the p-value
- Do bootstrapping with map
- Plot histogram, red line with null hypothesis
- Calculate p value from bootstrap hist
- Show side by side comparison of both p-values
- Use automated
moderndive::functions to get the bootstrapped p-value?
Summary
- Use
t.test()to do t-test in R - Using bootstraps/re-sampling distributions to get p-values and understand t-test
- Standard Error of Mean (SEM)
- Confidence intervals (CI)